AI + Biology: DeepMind releases massive protein structure database
- Determining a protein’s structure is crucial to understanding its function, but the process is long and difficult.
- Now, AI can predict a protein structure just based on its amino acid sequence with a high level of accuracy.
- The deputy director general of the European Molecular Biology Laboratory said, “This will be one of the most important datasets since the mapping of the Human Genome.”
This article was originally published by our sister site Freethink.
DeepMind, a sister company of Google, is giving the world access to a massive protein structure database — a gift that has the potential to revolutionize scientific research.
“This will be one of the most important datasets since the mapping of the Human Genome,” Ewan Birney, deputy director general of the European Molecular Biology Laboratory, which partnered with DeepMind on the database, said in a press release.
Protein structure: Proteins are molecules that are hugely important to the functioning of living organisms, including humans — practically everything we’re made of and everything our cells do is determined by our proteins.
“It’s the most significant contribution AI has made to advancing scientific knowledge to date.”
DEMIS HASSABIS
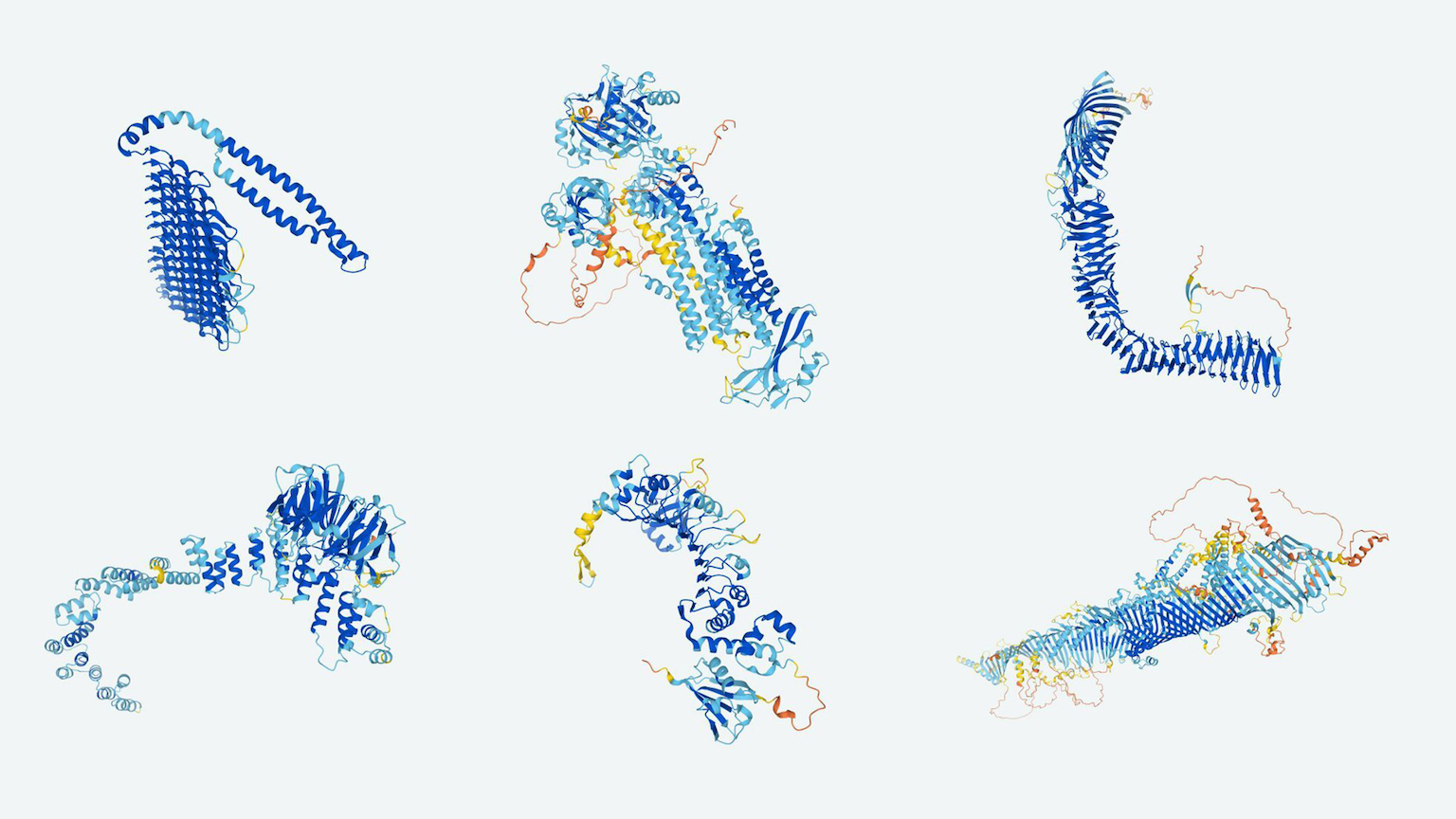
Every protein is made up of a long string of hundreds or even thousands of chemical compounds called amino acids, and the way that ribbon folds on itself determines the protein’s function.
Once scientists know a protein’s 3D structure, they know how it interacts with everything else, and they can start exploring ways to use the molecule to develop drugs, study diseases, design energy systems, and more.
The challenge: While it is possible for scientists to see protein structure, it isn’t easy — the standard method involves x-ray crystallography, which is about as expensive and complicated as it sounds.
The process isn’t fast, either. Determining a single protein structure can take anywhere from weeks to months, and after decades of work, scientists have only deciphered about 17% of the human body’s 20,000 proteins, known collectively as the proteome.
Christmas in July: In an attempt to solve science’s protein folding problem, DeepMind created AlphaFold, an AI that can predict a protein structure just based on its amino acids with a high level of accuracy in just a day or two.
Now, the company has announced that it’s making a database of the AI’s predictions freely available online.
Not only does this database contain structure predictions for all 20,000 proteins in the human body, it also includes 330,000 other proteins found in 20 organisms regularly used for scientific research, including mice, zebrafish, and fruit flies.
There’s more to come, too: DeepMind expects to release at least 100 million more protein structure predictions in the next few months. At that point, the database will include every protein known to science.
“We believe this represents the most significant contribution AI has made to advancing scientific knowledge to date, and is a great illustration of the sorts of benefits AI can bring to society,” DeepMind Founder and CEO Demis Hassabis said in the press release.